Research Outline
Our projects aim to understand the molecular mechanisms of
circadian rhythms in Arabidopsis and the role of
circadian regulation in plant physiology. Circadian rhythms
reflect the output of an intracellular circadian oscillator (or 'biological clock'). A
long-standing interest of ours is in the mechanism of this oscillator, which we're
investigating using clock mutants and circadian-regulated genes.
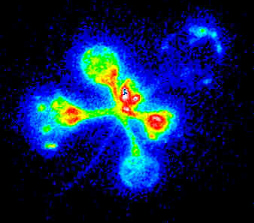
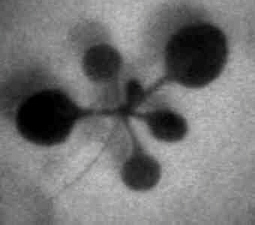
Results from an experiment...
Firefly luciferase (luc) activity in a transgenic Arabidopsis plant. The "normal" reference image on the right was taken with external lighting. The luminescence image on the left is the glow emitted by luciferase activity within the plant (on a computer-generated colour scale, with red indicating bright luminescence and blue low luminescence - none is visible to the naked eye).
The spatial and temporal pattern of luminescence reflects the expression pattern of the CAB gene promoter that controls luciferase in this transgenic plant. The highest levels are in young leaves, with little in the seedling stem (hypocotyl, extends to lower left).
Click here for a graph showing the circadian rhythm in luminescence from a wild-type CAB:luc seedling, and from a mutant (toc1-1). The mutant clock runs faster than normal. This was the first clock mutant identified in plants (Millar et al., Science, 1995). The Kay lab's cloning of the mutated gene has recently been reported (Strayer et al., Science, 2000). A growing collection of clock genes is revealing more of the important components of the plant clock.
Current projects
Our experimental methods combine real-time video imaging of transcription and signal transduction in transgenic plants carrying bioluminescent reporter genes (luciferases), with Arabidopsis thaliana mutants and molecular approaches, including microarray measurements of RNA levels. Our theoretical methods centre on differential equation models, parameter estimation and data analysis.
A control network within each cell. The plant cell integrates timing signals with a variety of other stimuli, to ensure appropriate responses to the environment throughout the day/night cycle. For example, many clock-regulated processes are also controlled by light signals from the environment, via plant photoreceptors. Our gating project studies how the cell balances light and timing information by rhythmically regulating its response to light. This mechanism of "cross-talk" between signalling pathways, known as circadian gating, is common to many organisms. In plants, it probably occurs within single cells. We are studying how the clock controls the expression and function of phytochrome photoreceptors. We showed that the gating mechanism affects light input to the clock - in fact, gating is essential for the clock to run under long days.
Modelling the clock. This collaborative project uses mathematical analysis and simulation to understand the plant clock mechanism, and by comparison to other clock models, to uncover the design principles underlying circadian clocks in all organisms. We now have a rich exchange between theory and experiment: improving our experimental designs through simulated experiments and validating mathematical models with real data.
A systems-oriented database of rhythm data.
Software for systems biology.
Past projects
Clocks under natural selection. Circadian clocks in all organisms are reset by environmental signals like the light and temperature changes at dawn and dusk. However, it's also important for clocks to keep accurate time under a range of environmental conditions. We're using quantitative genetics (QTL analysis) to compare the clocks of Northern European and subtropical Arabidopsis strains and find clock genes that differ between the strains. These variant genes may have been retained during recent evolution of the clock in the very different native habitats of the strains. We identified new clock-associated genes in this way, and also studied which aspects of the clock have been modified to allow Arabidopsis to adapt to new environments (Edwards et al. 2005, Genetics). Similar modifications could also extend the range of agriculturally important plants, such as Brassica oleracea.
Autonomy or cooperation? Many or all plant cells contain a clock and photoreceptors. Some animal cell clocks are linked to each other or to a central pacemaker in the brain. Our cell-autonomy project aimed to determine whether plant cells compare time and lighting information with their neighbours and whether plants have a central circadian pacemaker. We have shown that each plant organ contains at least one circadian system that is independent in the intact plant: plants have no pacemaker (Thain et al. 2000, Current Biology). Rhythms in epidermal cells can spontaneously desynchronise from rhythms in the underlying leaf mesophyll cells, indicating that clocks are not only independent but can also differ among cell types.
| Arabidopsis thaliana | |
| Circadian Rhythms | |
| Video Imaging | |
| Luciferase | |
| Rhythm movies |
![]()