New Models Download Page
Models for Circadian Modelling Version 2.0
Click on the links below to download a model zip file. Follow the instructions below to install the model into Circadian Modelling version 2.0.
- Arabidopsis Models
- J. C. W. Locke, A. J. Millar & M. S. Turner (2005a). Modelling genetic networks with noisy and varied experimental data: the circadian clock in Arabidopsis thaliana. J. theor. Biol. 234(3): 383-393 Download
- J. C. W. Locke, M. M. Southern, L. Kozma-Bognar, V. Hibberd, P. E. Brown, M. S. Turner, A. J. Millar (2005b). Extension of a genetic network model by iterative experimentation and mathematical analysis. Molecular Systems Biology. 1:13 Download. Also available from BioModels as BIOMD0000000055.
- Locke JCW, Kozma-Bognar L, Gould PD, Fehér B, Kevei E, Nagy F, Turner MS, Hall A, Millar AJ (2006) Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana.. Molecular Systems Biology. 2:59 Download. Also available from BioModels on the Uncurated models page.
- Neurospora Models
- J.-C. Leloup & A. Goldbeter (1999). Limit cycle models for circadian rhythms based on transcriptional regulation in Drosophila and Neurospora. J. Biol. Rhythms 14(6):433-48 Download
- Mammalian Models
- Drosophila models
- R. Ueda, M. Hagiwara & H. Kitano. (2001). Robust oscillations within the interlocked feedback model of Drosophila circadian rhythm. J. theor. Biol. 210:401-406 Abstract Download
- J.-C. Leloup & A. Goldbeter (1999). Limit cycle models for circadian rhythms based on transcriptional regulation in Drosophila and Neurospora. J. Biol. Rhythms 14(6):433-48 Download
Given the spread of tools for SBML, we aim to adapt our simulation tools to use SBML-compliant models rather than to build many more models for CM.
Models for Circadian Modelling Version 1.x
Click on the links below to download a model zip file. Follow the instructions below to install the model into Circadian Modelling version 1.x.
- Arabidopsis Models
- J. C. W. Locke, A. J. Millar & M. S. Turner (2005a). Modelling genetic networks with noisy and varied experimental data: the circadian clock in Arabidopsis thaliana. J. theor. Biol. 234(3): 383-393 Download
- J. C. W. Locke, M. M. Southern, L. Kozma-Bognar, V. Hibberd, P. E. Brown, M. S. Turner, A. J. Millar (2005b). Extension of a genetic network model by iterative experimentation and mathematical analysis. Molecular Systems Biology. In press Download
- Mammalian Models
- D. B. Forger & C. S. Peskin (2003). A detailed predictive model of the mammalian circadian clock. PNAS 100(25): 14806-14811 Abstract
- J.-C. Leloup & A. Goldbeter (2003). Toward a detailed computational model for the mammalian circadian clock. PNAS 100(12): 7051-7056 Abstract
- Version b. This is the 19 equation version which contains the Rev-Erb-α gene
- Drosophila models
- R. Ueda, M. Hagiwara & H. Kitano. (2001). Robust oscillations within the interlocked feedback model of Drosophila circadian rhythm. J. theor. Biol. 210:401-406 Abstract Download
- J.-C. Leloup & A. Goldbeter (1999). Limit cycle models for circadian rhythms based on transcriptional regulation in Drosophila and Neurospora. J. Biol. Rhythms 14(6):433-48 Download
- Neurospora Models
- J.-C. Leloup & A. Goldbeter (1999). Limit cycle models for circadian rhythms based on transcriptional regulation in Drosophila and Neurospora. J. Biol. Rhythms 14(6):433-48 Download
Installing a new model
- Extract the model files from the zip file to a suitable folder
- If the model is a replacement for a previous version with the same name and is to be installed in the same model group, then the original model should be completely removed first.
- This is done via
the
Model->Uninstallmenu item. Ensure that the folder<INSTALL_DIR>\Models\Group Name\Model Namehas been deleted.<INSTALL_DIR>is the folder in which Circadian Modelling is installed,Group Nameis the name of the model group, andModel Namethe name of the model. Note that although you will be given an opportunity to save any results files for the model before deletion, any parameters, environment or mutations files will be lost, unless you back up the folders<INSTALL_DIR>\Models\Group Name\Model Name\Parameters, <INSTALL_DIR>\Models\Group Name\Model Name\Environment,and<INSTALL_DIR>\Models\Group Name\Model Name\Mutationsmanually. This may be worth doing if the model itself is identical to the older version, with only the web components being updated. This is the case with the Leloup & Goldbeter model Version b above. This means that any parameters, environment or mutations files created with the version on our original distribution discs will be fully compatible with this new version.
- To install a new model select the
Model->Installmenu item, or click the corresponding toolbar button . You will then be prompted to locate the executable file, extracted from the
zip file, representing the model. All other required files are expected to be found in the same folder
in order for the installation to proceed correctly.
. You will then be prompted to locate the executable file, extracted from the
zip file, representing the model. All other required files are expected to be found in the same folder
in order for the installation to proceed correctly. - Next, you will be asked to specify a group to which the new model is to belong.
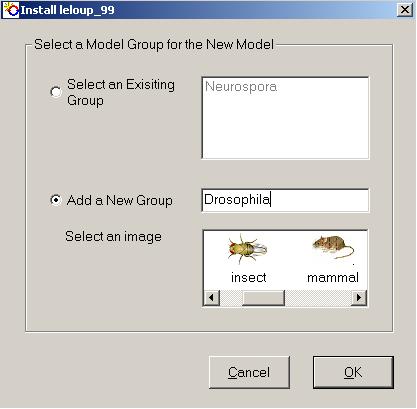
- By selecting the appropriate option button, a pre-existing group can be selected from the list, or a new group can be created. The purpose of grouping the models is to make them easier to manage and to locate the desired one. This will be especially important when a large number are installed. Another advantage is that, although all models within a group must have unique names, multiple models with the same name can exist in different groups.
- When creating a new group, you can select an image to represent it. This will be the image appearing next to the group name in the lefthand list box on the Circadian Modelling main window. It is assumed that most users will want to group models according to organism, hence a number of images are provided representing the model organisms commonly used in circadian biology. However, groups can take any name at all and some users may wish to organise their collections differently, for example, grouping models by author. This will make no difference at all to the program.
- Installation should then be completed automatically and the new model is then immediately available for selection.